Activity-Based Biochemical Phosphatase Panel Screening and Profiling Services
Real-Time Functional Insights for Phosphatase-Targeted Drug Discovery
AssayQuant’s KinSight™ Phosphatase Profiling Platform provides continuous, real-time enzymatic activity measurements for a growing panel of serine/threonine and tyrosine phosphatases. Unlike traditional phosphatase services that rely on DiFMUP endpoint assays, we use physiologically relevant phosphopeptide substrates and kinetic readouts to reveal true potency, selectivity, and mechanism of action (MOA).
If you’re advancing phosphatase inhibitors, validating targets, or improving SAR, our kinetic platform delivers the actionable insight traditional assays miss.
When you’re ready for high-throughput screening (HTS), the same assay chemistry seamlessly converts into our PhosphoSens-Red TRF endpoint format — adding Europium for a 100 µsec delay and 620 nm emission to eliminate compound autofluorescence and support large-library screening.
Go Beyond % Inhibition: Get Actionable Kinetic Insights with KinSight™ Phosphatase Profiling
True Kinetic Understanding:
Directly measure activity and quantify % inhibition - no endpoint assumptions - for accurate potency, selectivity, and MOA insights.
Optimized Peptide Substrates :
Designed around native motifs, avoiding limitations of DiFMUP assays
Actionable Reports, Fast:
Receive comprehensive reports with expert annotations highlighting exploitable kinetics (like TDI) – all within two weeks.
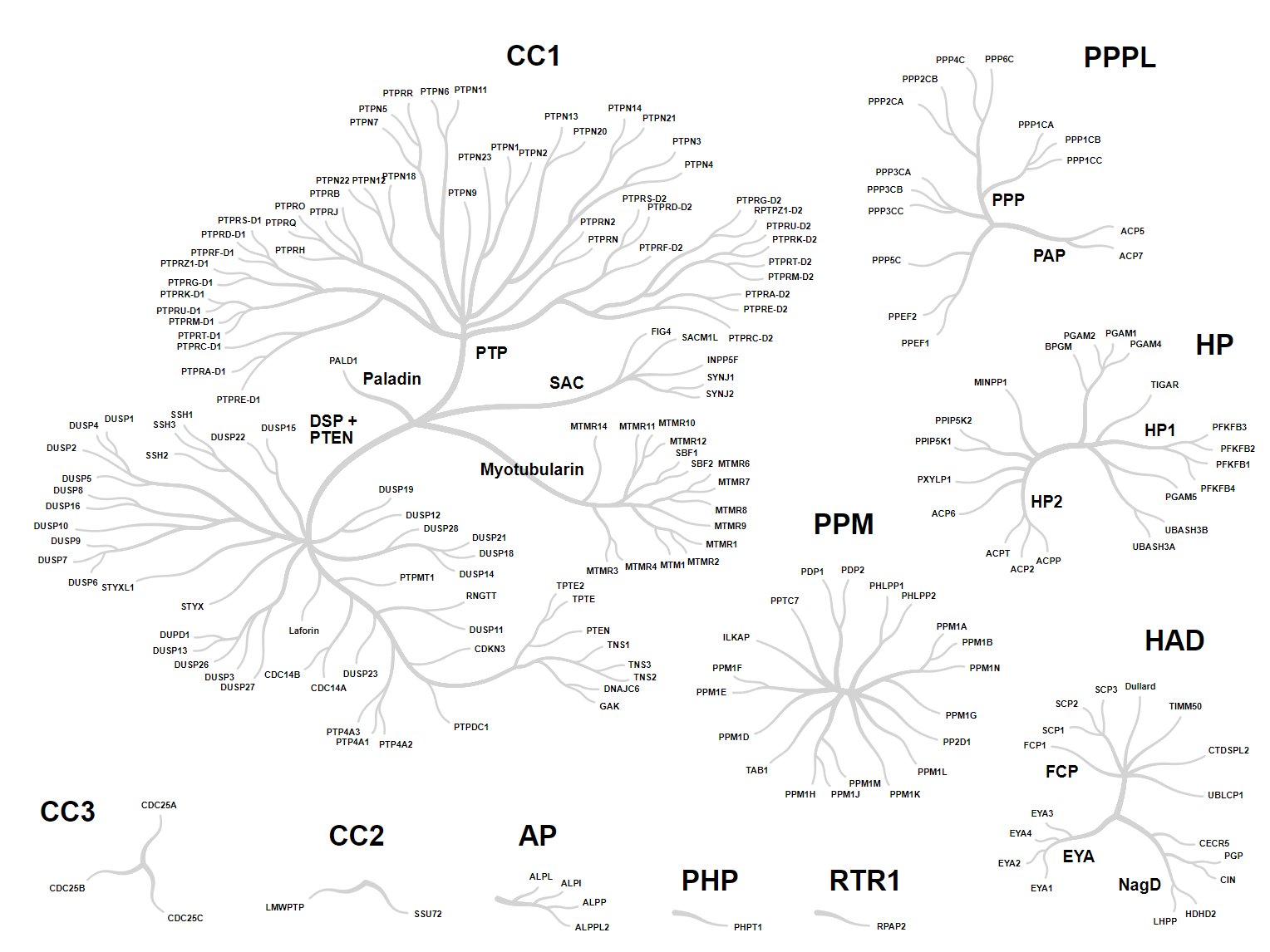
Phosphatase family tree generated using CoralP.
Phosphatase Selectivity Panels
Historically viewed as difficult drug targets, phosphatases are experiencing renewed interest — with major advances in tyrosine and serine/threonine phosphatase-targeted therapies driving fresh momentum in the field.
We provide biochemical profiling across key phosphatase families using real-time kinetic assays. Each study delivers deep functional insight into selectivity, potency, and inhibitory mechanism.
Testing Performed
Phosphatase selectivity profiling using our PhosphoSens Technology generates a progress curve in everywell, providing you with actionable data you can be confident in. Selectivity screening available :
- Across a variety of predefined and customizable panels
- Single compound concentration or multiple doses available
- All performed in duplicate, n=2
Swiftly determine phosphatome-wide selectivity, gain crucial insights into mechanism of action and relative potency directly from the profiling data.
Deliverables
Detailed reports for each compound designed to guide your development decisions with annotated progress curves to call out exploitable mechanisms of actions (MOA) and % inhibition presented in:
- Numerical Format (Tabular)
- Graphical Format (Waterfall Plot)
- Visual Format (Phosphatase Tree)
- Progress Curves
Screening complete in two weeks so you get results and important functional information about the enzymology and nature of the compound quickly!
34 Protein Phosphatases Profiled |
|---|
| Protein Tyrosine Phosphatase, CDC25A |
| Protein Tyrosine Phosphatase, CDC25B |
| Protein Tyrosine Phosphatase, CDC25C |
| Protein Tyrosine Phosphatase, DUSP22 (JSP-1, LMW-DSP2, MKPX) |
| Protein Tyrosine Phosphatase, DUSP3 (VHR) |
| Protein Serine/Threonine Phosphatase, PPM1A (PP2Ca, PP2CA) |
| Protein Serine/Threonine Phosphatase, PPM1D (WIP1, PP2C-delta) |
| Protein Serine/Threonine Phosphatase, PPM1G (PP2Cg) |
| Protein Serine/Threonine Phosphatase, PPM1F (CamKPase) |
| Protein Serine/Threonine Phosphatase, PPP1CA (PP1A/PPP1A) |
| Protein Serine/Threonine Phosphatase, PPP1CB (PP1B, PPP1B) |
| Protein Serine/Threonine Phosphatase, PPP2CA (PP2A-α, RP-C) |
| Protein Serine/Threonine Phosphatase, PPP2CB (PP2Abeta) |
| Protein Serine/Threonine Phosphatase, PPP3CA/B (Calcineurin A/B) |
| Protein Tyrosine Phosphatase, PTPN1 (PTP1B, PTB1B) |
| Protein Tyrosine Phosphatase, PTPN2 (TCPTP, PTPT) |
| Protein Tyrosine Phosphatase, PTPN4 (MEG1) |
| Protein Tyrosine Phosphatase, PTPN5 (STEP) |
| Protein Tyrosine Phosphatase, PTPN6 (SHP-1, PTP-1C) |
| Protein Tyrosine Phosphatase, PTPN7 (HEPTP, LC-PTP) |
| Protein Tyrosine Phosphatase, PTPN9 (MEG2) |
| Protein Tyrosine Phosphatase, PTPN11 (SHP-2, PTP2C, PTP1D) |
| Protein Tyrosine Phosphatase, PTPN12 (PTP-PEST, PTPG1) |
| Protein Tyrosine Phosphatase, PTPN13 (FAP-1, PTP-BAS, PTP-E1, PNP1) |
| Protein Tyrosine Phosphatase, PTPN22 (PTPN8, Lyp) |
| Protein Tyrosine Phosphatase, PTPRA (R-PTP-alpha, PTPA, LRP) |
Additional targets can be added upon request
Stop Using Outdated Assays—Upgrade to KinSight Today
AssayQuant's KinSight™ profiling services leverage our unique PhosphoSens® Technology, a continuous assay format that directly measures substrate phosphorylation throughout the entire reaction. Instead of relying on single-point estimates, this approach generates a full progress curve in every well, yielding the actual reaction rate from dozens of data points.
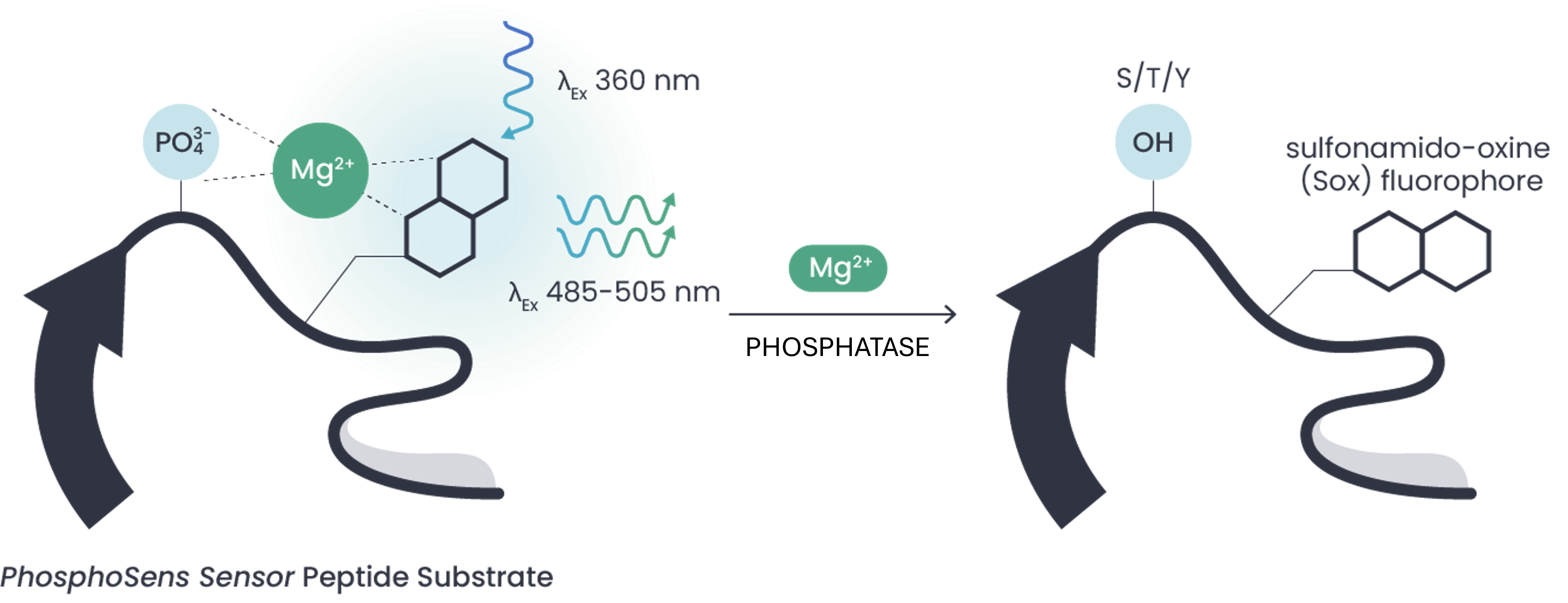
Understanding the true reaction rate provides higher confidence data, free from the assumptions inherent in endpoint assays. This makes KinSight™ profiling valuable at all stages of preclinical drug discovery, helping identify compounds with poor selectivity, pinpoint optimization opportunities, or confirm off-target interactions early.
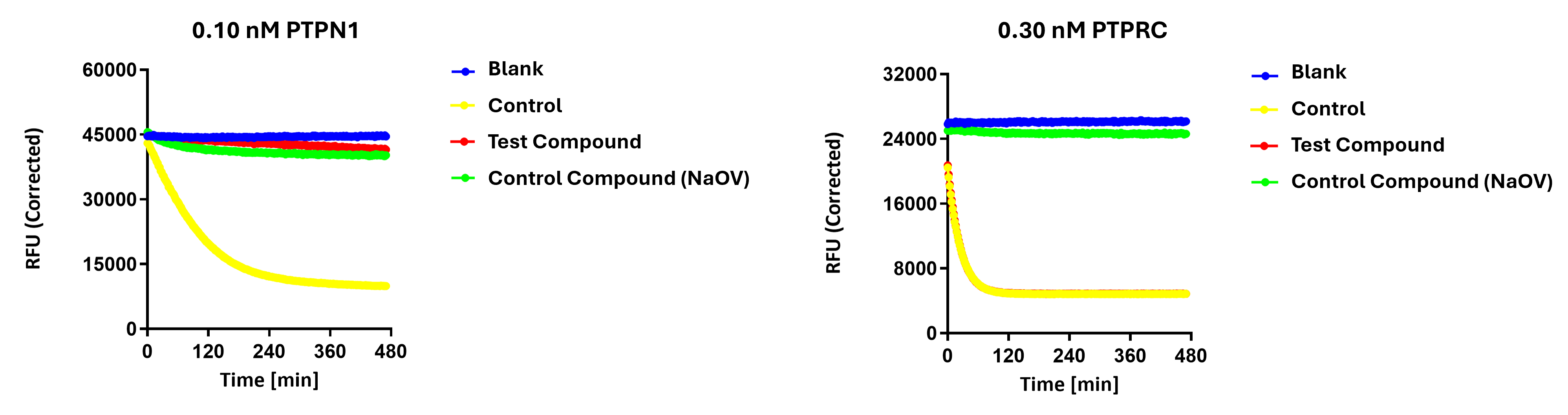
Why Most Phosphatase Profiling Falls Short
Most commercial phosphatase profiling services use DiFMUP, a generic fluorogenic reporter substrate. While convenient, DiFMUP introduces several scientific limitations that reduce biological relevance and obscure true inhibitor behavior.
Limitations of DiFMUP-Based Phosphatase Assays
| Limitation | Why It Matters |
|---|---|
| Generic, non-physiological substrate | Does not reflect native phosphosite sequence or charge → leads to misleading potency and missed specificity. |
| Single time-point endpoint readout | Cannot capture TDI, reversible vs irreversible inhibition, or kinetic behavior across time. |
| High fluorescence interference | DiFMUP’s phenolic fluorophore overlaps with many small molecules → increased false positives. |
| Overestimates inhibitor potency | Artificial substrate hydrolysis inflates IC₅₀ values, making SAR misleading. |
| Cannot resolve substrate-specific selectivity | Phosphatases recognize native sequence motifs—DiFMUP masks true biological selectivity. |
How PhosphoSens Solves These Issues
- Optimized phosphopeptide substrates rationally designed from native phosphosite motifs
- Real-time continuous kinetics instead of single-point data
Bottom line: DiFMUP tells you whether inhibition occurs. PhosphoSens tells you how fast, how strong, and on which physiologically relevant substrate.
The KinSight Advantage: Real-Time Kinetic Data
Powerful Kinetic Assays Enabling Protein Phosphatase Drug Discovery
Protein phosphatases are an important drug target in the development of cancer therapies as the overexpression of these proteins can activate signaling pathways and promote tumor development.
While phosphatases were once considered a challenging drug target, new advancements have created a renewed focus on tyrosine phosphatase-targeting therapies. Development of these drug candidates creates a need for robust assays that monitor phosphatase enzyme activity.
Harnessing our PhosphoSens® technology using novel Sox-based phosphopeptide substrates, we developed assays to understand protein phosphatase activity. Download this poster to learn more today:
Ready to Profile Your Compounds?
Phosphatases are complex, highly regulated enzymes. A single endpoint doesn’t tell the story.
Real-time kinetic data does.
Resources
AssayQuant's Kinome Profiling Services Are More Than Just an Alternative—Access A Better Approach to Kinome Profiling Today
Most biochemical kinase profiling services rely on endpoint assays, providing only a single snapshot of enzyme activity. But kinase inhibition is dynamic—why settle for incomplete data?
AssayQuant's KinSight Kinase Panel Screening and Profiling Services deliver everything you need—and more—with a next-generation approach to kinase inhibitor characterization.
With AssayQuant's KinSight profiling platform, you don’t just get data—you get kinetic insights into selectivity and mechanism of action (MOA) for your drug discovery research.
Kinome Tree Mapping Application
Visualize your kinome profiling data with customizable kinome trees.
Kinome Profiling With a Continuous Assay
Stay Informed
Want to hear the latest about our technology? Be among the first to learn about our latest products and services.